↗ OpenReview ↗ NeurIPS Proc. ↗ Hugging Face ↗ Chat
TL;DR#
Volumetric medical image segmentation is crucial for diagnosis and treatment planning but faces challenges like limited training data and computationally expensive inference. Existing methods often struggle with complex structures and lack user-friendly interaction. This necessitates a universal model capable of handling various anatomical structures efficiently.
SegVol addresses these challenges with a novel 3D foundation model trained on a massive dataset. It incorporates a zoom-out-zoom-in mechanism to optimize inference and allows user interaction via spatial and semantic prompts. SegVol outperforms current state-of-the-art methods on 22 anatomical segmentation tasks, demonstrating its effectiveness and potential to advance volumetric medical image analysis.
Key Takeaways#
Why does it matter?#
This paper is crucial for researchers in medical image analysis because it introduces SegVol, a groundbreaking 3D foundation model for universal and interactive medical image segmentation. SegVol’s ability to handle diverse anatomical categories and user interactions opens exciting new avenues for research and development. Its impact extends to various clinical applications, including tumor monitoring and surgical planning, making it highly relevant to current trends in AI-driven healthcare.
Visual Insights#

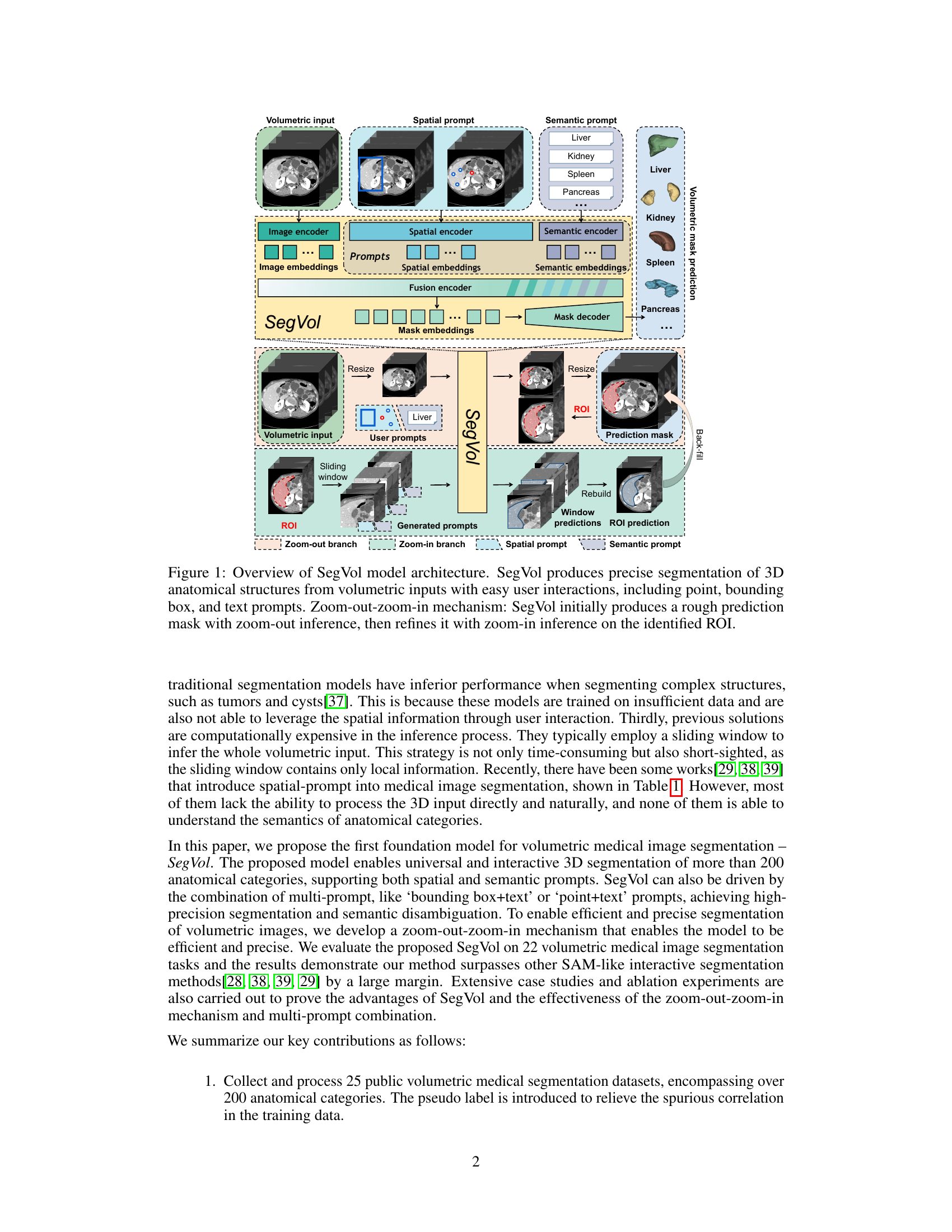
This figure illustrates the architecture of the SegVol model, which is designed for universal and interactive volumetric medical image segmentation. The model takes volumetric medical images as input, along with user-provided prompts (point, bounding box, or text). It utilizes a zoom-out-zoom-in mechanism to first generate a rough prediction mask using a reduced resolution image. This rough prediction is then used to identify a Region of Interest (ROI), which is further processed at a higher resolution to refine the segmentation mask. The overall workflow involves several encoders (image, spatial, and semantic) and a decoder to produce the final volumetric mask.

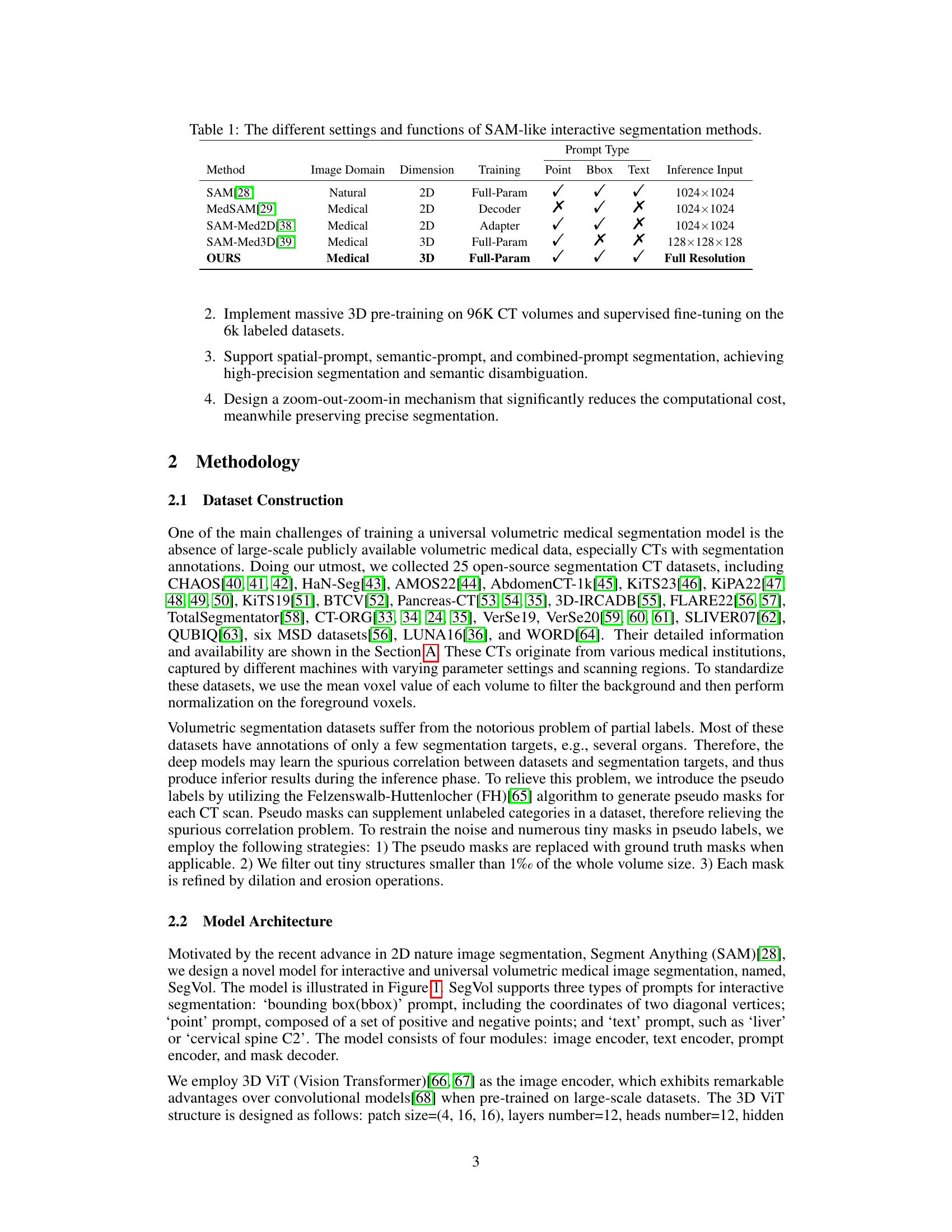
This table compares several state-of-the-art interactive segmentation methods similar to Segment Anything (SAM). It contrasts their image domain (natural vs. medical), dimensionality (2D vs. 3D), training approach (full parameter training, decoder only, adapter), and the types of prompts they support (point, bounding box, text). Finally, it shows the input resolution used during inference.
In-depth insights#
3D Seg. Foundation#
A 3D segmentation foundation model is a significant advancement in medical image analysis. Universality across diverse anatomical structures is key, enabling applications beyond current task-specific models. The approach of scaling training data to include many unlabeled volumes is crucial for achieving robustness and generalization. Interactive segmentation through semantic and spatial prompts allows for user input and refinement of predictions, which is essential for clinical use. The efficiency of the inference process via a zoom-out/zoom-in mechanism is vital to make the model practical for real-world applications. High precision compared to existing methods and ability to resolve ambiguities via combined prompts are significant strengths. Future work should focus on expanding the model to encompass additional modalities and address the challenges of segmenting complex structures and handling spurious correlations in datasets.
Zoom-Out/In Inference#
The proposed “Zoom-Out/Zoom-In Inference” strategy is a computationally efficient approach to volumetric medical image segmentation. It cleverly addresses the challenge of processing large 3D images by first performing a coarse, zoom-out segmentation to identify regions of interest (ROIs). This initial pass provides a rough prediction mask. Subsequently, the model focuses on the identified ROIs with zoom-in inference, allowing for higher-resolution, more precise segmentation within these areas. This two-stage process avoids the computational burden of processing the entire volume at full resolution, making it suitable for real-world applications. The use of multi-size training further enhances the efficiency and accuracy of the model. This process also offers a user-friendly interaction, as the initial low-resolution prediction helps to guide subsequent interactions with the high-resolution processing for specific ROIs.
Multi-Prompt Fusion#
Multi-prompt fusion is a crucial technique in leveraging the power of diverse input modalities for enhanced performance. By combining spatial prompts (e.g., bounding boxes, points), which provide precise location information, with semantic prompts (e.g., text descriptions), which offer contextual understanding, a model can achieve significantly improved accuracy and robustness. The fusion process needs careful design, as simply concatenating embeddings from different sources may not capture their intricate relationships. Effective fusion strategies might involve attention mechanisms to weigh the importance of each prompt based on the specific context or feature interaction, or transformer-based architectures allowing rich cross-modal communication. The success of multi-prompt fusion highly depends on the quality and diversity of training data. A model trained on a wide variety of cases and prompt combinations will naturally exhibit better generalization capabilities. Furthermore, the ability to handle missing or incomplete prompts is a critical aspect, as in real-world scenarios, complete information is not always available. Robust fusion methods should incorporate strategies for gracefully handling missing inputs without substantially degrading performance.
Ablation Studies#
Ablation studies systematically investigate the contribution of individual components within a machine learning model. In the context of a volumetric medical image segmentation model, this could involve removing or modifying different modules (e.g., the image encoder, the prompt encoder, the mask decoder) to assess their impact on overall segmentation accuracy and efficiency. Key aspects explored might include the effectiveness of the zoom-out-zoom-in mechanism, which likely balances computational cost with segmentation precision. The influence of various prompt types (spatial, semantic, or combined) would also be examined, revealing whether specific prompts are crucial for certain anatomical structures or tasks. Results would demonstrate the importance of specific design choices, quantifying their contribution to the final performance. Additionally, the ablation study would likely analyze the impact of dataset size, revealing whether increasing the quantity of training data disproportionately improves segmentation performance.
Future of SegVol#
SegVol’s future hinges on several key areas. Expanding its capabilities to encompass additional medical imaging modalities beyond CT scans (e.g., MRI, ultrasound) is crucial for broader applicability. Improving its efficiency and reducing computational costs, especially for high-resolution 3D images, will enhance real-world usability. Integration with existing clinical workflows through seamless API access and compatibility with standard medical imaging platforms is essential for mainstream adoption. Addressing the limitations of relying on pseudo-labels and enhancing the model’s ability to handle noisy or incomplete data are vital for robustness. Finally, incorporating feedback mechanisms for continuous learning and adaptation would make the model more responsive and tailored to individual clinical needs. Exploring multi-modal learning, where SegVol combines information from various imaging modalities and clinical data, could unlock further advancements in diagnostic accuracy and personalized medicine. Furthermore, research into the model’s explainability and interpretability will greatly boost its trustworthiness and adoption by clinicians.
More visual insights#
More on figures

This figure presents a comparison of the performance of SegVol against other similar interactive segmentation methods across various datasets and anatomical structures. Violin plots visually represent the distribution of Dice scores for each method on each task, allowing for a comparison of both the central tendency and variability of performance. The x-axis indicates the different segmentation tasks (e.g., specific organs or lesions), and the y-axis represents the Dice score, a common metric for evaluating the accuracy of segmentation. The plots highlight SegVol’s superior performance in the majority of the tasks.

This figure provides a detailed overview of the SegVol model architecture, highlighting its key components and workflow. SegVol takes volumetric medical images as input and uses various types of prompts (point, bounding box, text) for interactive segmentation. The model’s core consists of an image encoder, spatial and semantic encoders, and a mask decoder. A notable feature is the zoom-out-zoom-in mechanism, where a rough prediction is generated initially at a lower resolution (zoom-out) before being refined with high-resolution processing (zoom-in) focused on the region of interest (ROI).

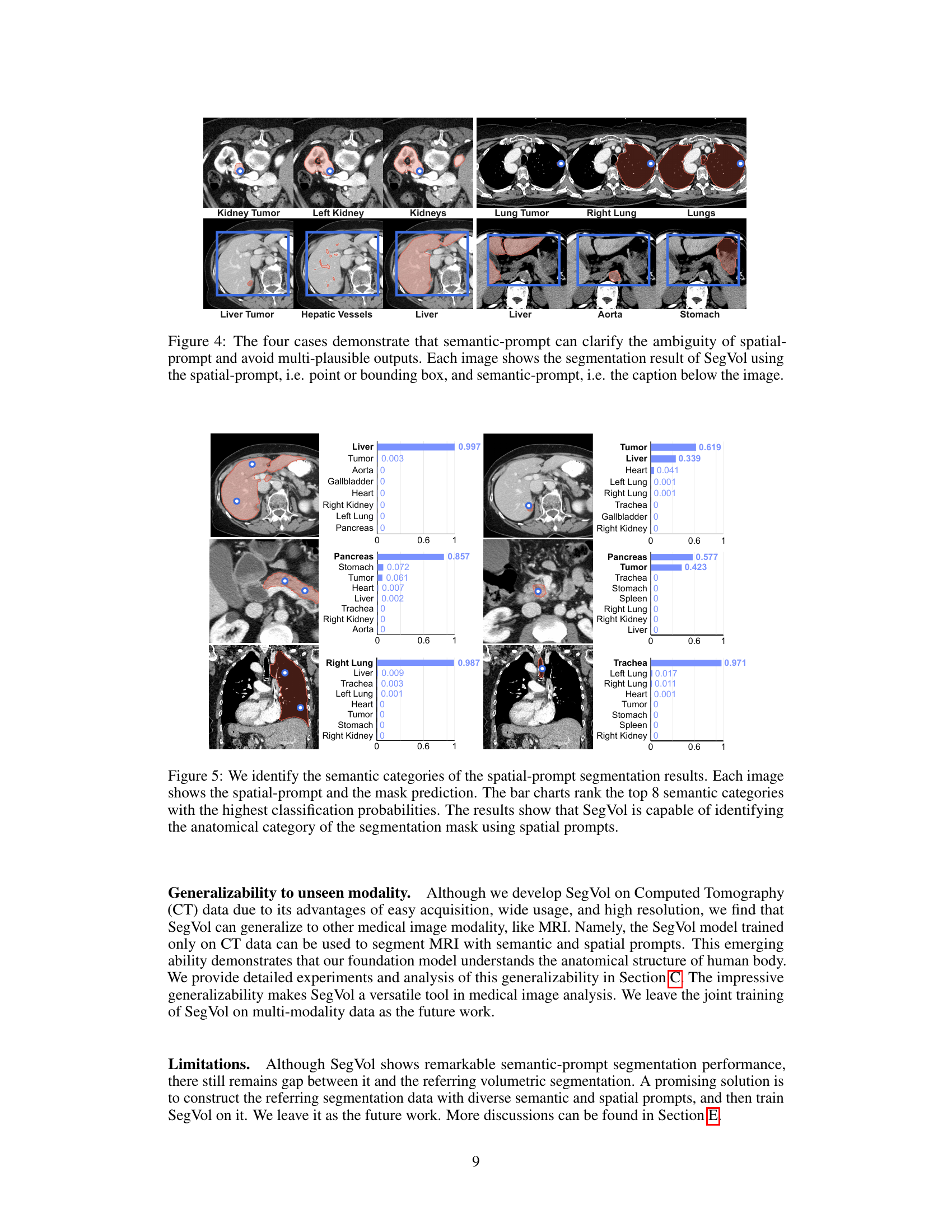
This figure shows four examples where a single spatial prompt (point or bounding box) could correspond to multiple anatomical structures. The ambiguity is resolved by adding a semantic prompt (text description) which specifies the target structure. SegVol then correctly segments the intended anatomy.

This figure provides a high-level overview of the SegVol model architecture, which is designed for universal and interactive volumetric medical image segmentation. It illustrates the model’s key components, including the image encoder, spatial and semantic encoders, fusion encoder, and mask decoder. The process starts with a volumetric input image and user prompts (point, bounding box, or text). The zoom-out-zoom-in mechanism is highlighted, showing how the model first generates a rough prediction mask using zoom-out inference and then refines it using zoom-in inference on the region of interest (ROI).

This figure illustrates the architecture of the SegVol model, a 3D foundation model for volumetric medical image segmentation. It shows the model’s components, including image, spatial, and semantic encoders, a fusion encoder, and a mask decoder. The figure also highlights the model’s interactive capabilities, supporting point, bounding box, and text prompts for user input. A key feature is the zoom-out-zoom-in mechanism, which allows for efficient and accurate segmentation by first generating a rough prediction mask and then refining it with a zoom-in step on the region of interest (ROI).

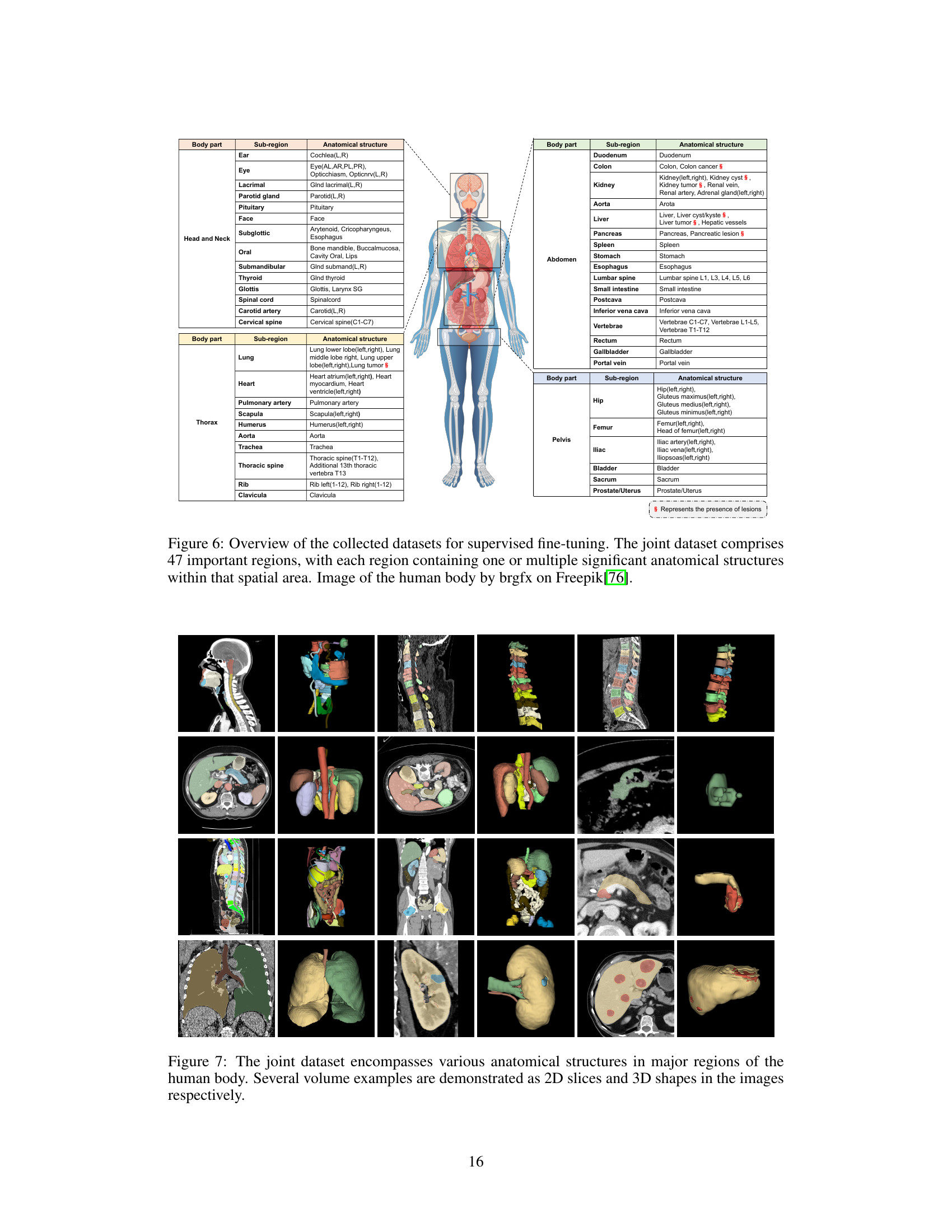
This figure shows examples of the various anatomical structures included in the joint dataset used to train the SegVol model. The images depict different body regions (head and neck, abdomen, thorax, pelvis) and demonstrate the range of anatomical structures included in the dataset, both as 2D slices and 3D renderings. This visualization helps to illustrate the diversity and comprehensiveness of the dataset, highlighting the complexity of volumetric medical image segmentation.
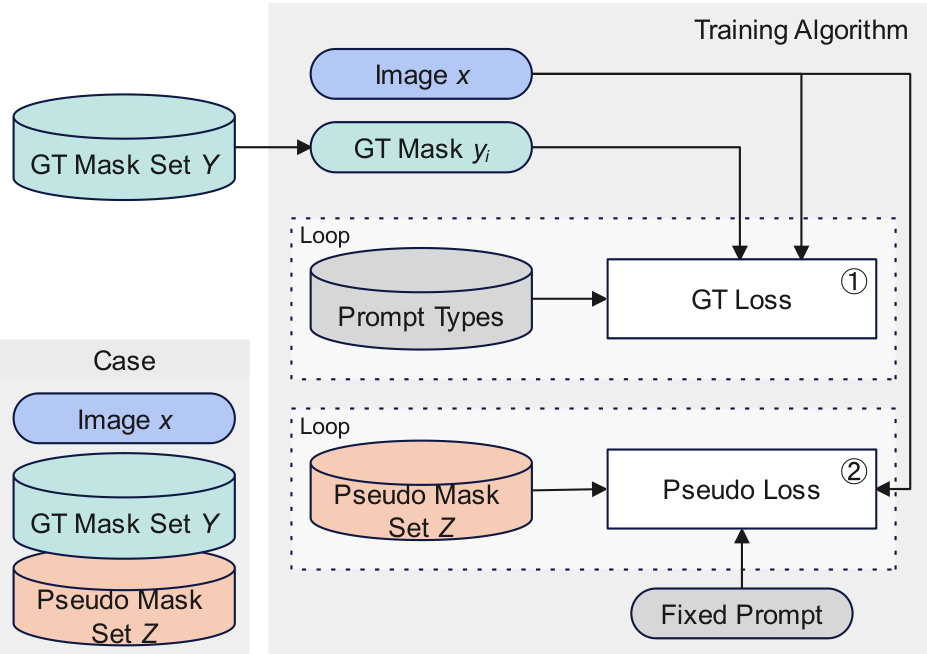
This figure illustrates the training algorithm of SegVol. Each training sample includes an image, a ground truth mask set, and a pseudo mask set. The training process involves two loops: one for the ground truth masks and one for the pseudo masks. Different prompt types are used in each loop. The ground truth loss and the pseudo loss are calculated separately and then combined to update the model parameters.
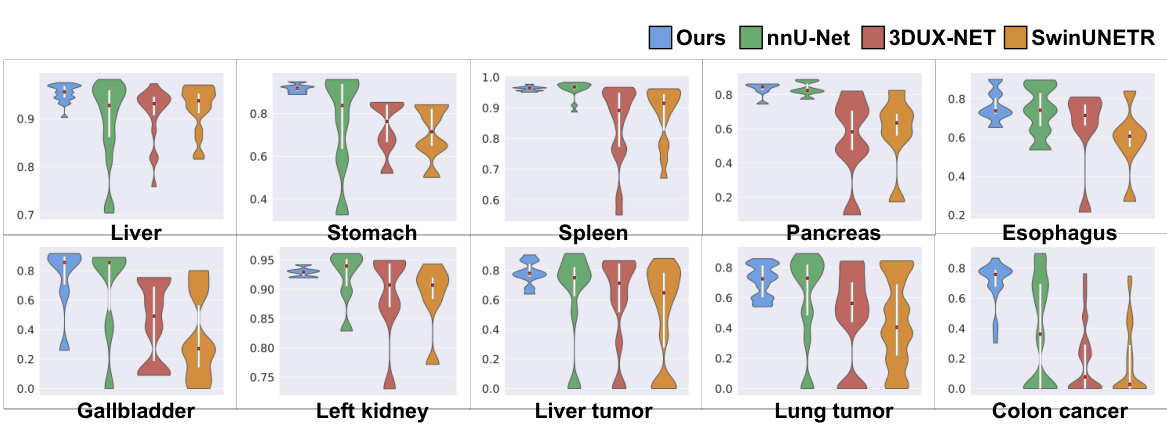
The figure shows a comparison of the performance of SegVol against three other task-specific methods (nnU-Net, 3DUX-NET, and SwinUNETR) across ten different organ and lesion segmentation tasks. Violin plots illustrate the distribution of Dice scores for each method and task, allowing for a visual comparison of performance variability and central tendency. The vertical axis represents the Dice score, a common metric for evaluating segmentation accuracy.
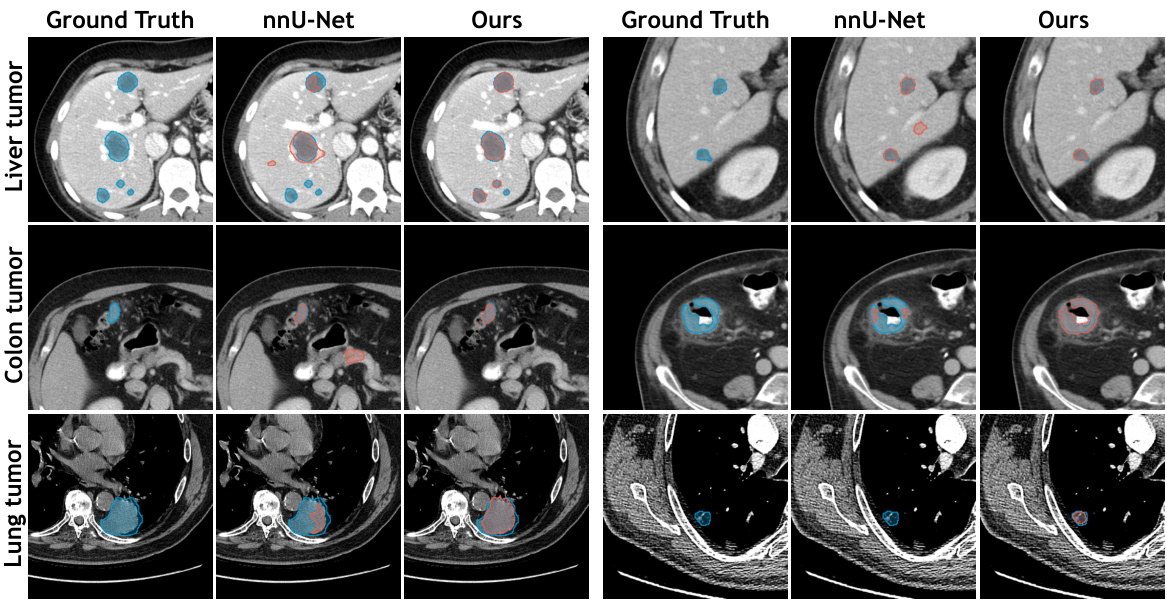
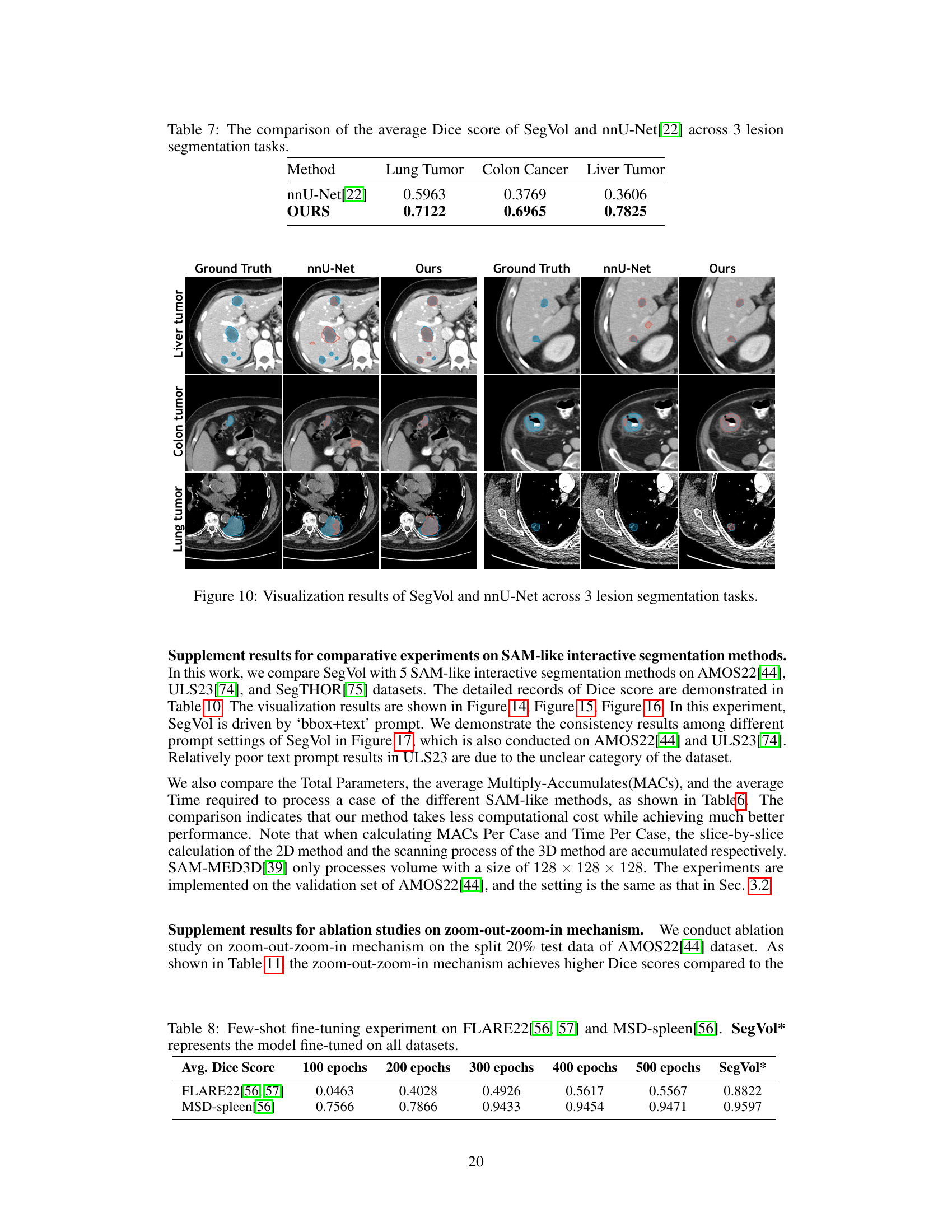
This figure visualizes the segmentation results of SegVol and nnU-Net on three lesion segmentation tasks: liver tumor, colon tumor, and lung tumor. For each task, it presents four image columns: the ground truth, the nnU-Net prediction, and the SegVol prediction. The visualization is meant to show a qualitative comparison of the segmentation performance of the two methods. The cyan (light blue) outlines represent the SegVol predictions while the red outlines represent the nnU-Net predictions. The goal is to allow visual inspection of the accuracy and precision of both methods.
This figure illustrates the architecture of the SegVol model, a 3D foundation model for universal and interactive volumetric medical image segmentation. It shows the process of taking a volumetric input and using various prompts (points, bounding boxes, text) to produce a precise segmentation mask. The key innovation is a zoom-out-zoom-in mechanism which starts by generating a rough prediction and then refines this prediction at a higher resolution on a region of interest (ROI).

This figure provides a detailed overview of the SegVol model architecture. It shows the different components of the model, including the image encoder, spatial encoder, semantic encoder, fusion encoder, and mask decoder. The figure also illustrates the zoom-out-zoom-in mechanism used for efficient and precise inference on volumetric images. This mechanism involves initially generating a rough prediction mask using zoom-out inference, then refining this mask using zoom-in inference on a smaller region of interest (ROI) identified from the initial prediction. The various types of user interactions supported by the model, including point, bounding box, and text prompts, are also shown.
This figure provides a detailed overview of the SegVol model architecture, illustrating its workflow for universal and interactive volumetric medical image segmentation. It highlights the key components, including the image encoder, spatial and semantic encoders, fusion encoder, mask decoder, and the zoom-out-zoom-in mechanism. The zoom-out-zoom-in process is shown to efficiently generate a precise segmentation mask by first creating a rough prediction and then refining it within the region of interest. The figure showcases how various user prompts (points, bounding boxes, text) are incorporated into the process for flexible interaction.

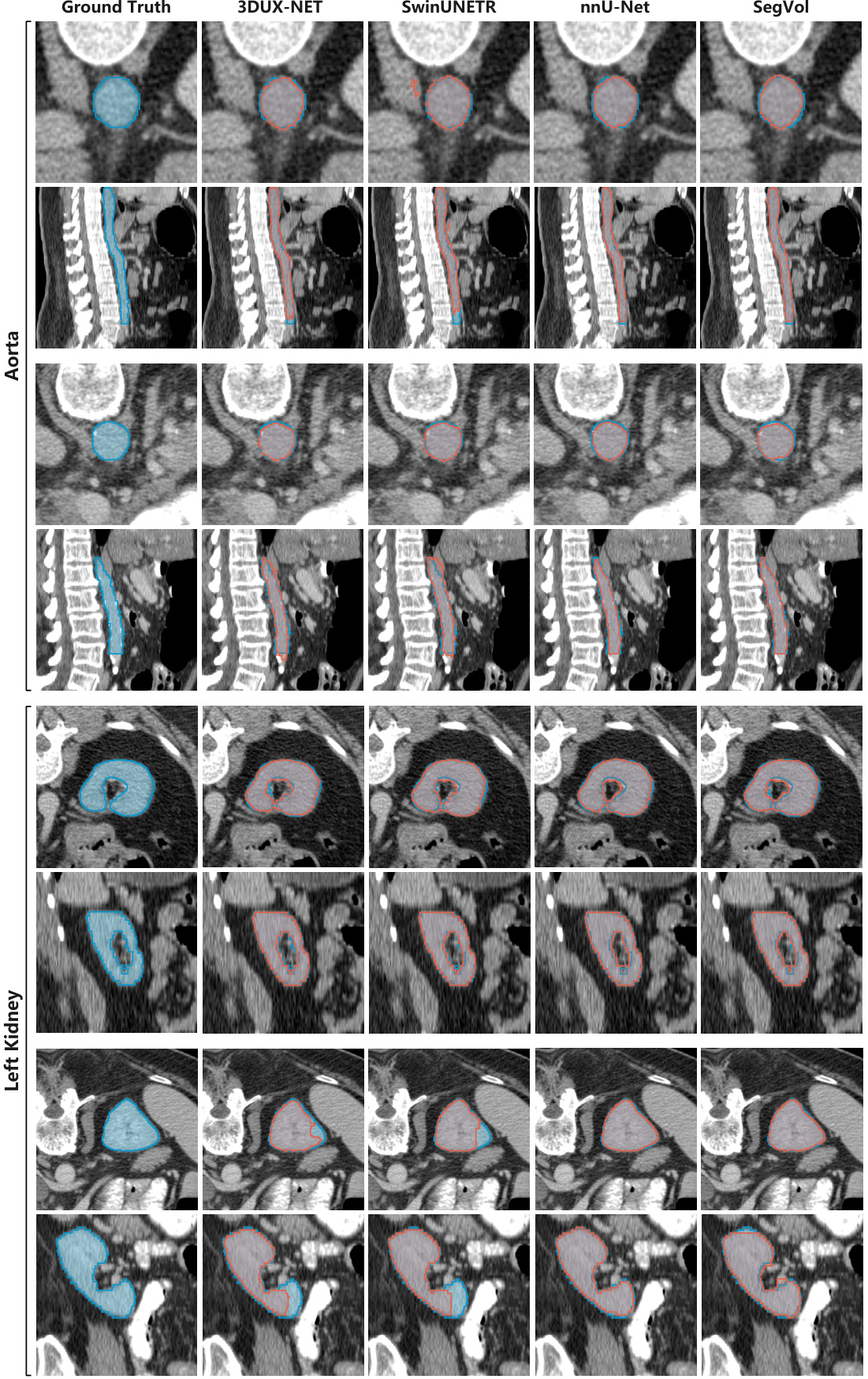
This figure compares the performance of SegVol against several other state-of-the-art interactive segmentation models, specifically MedSAM, SAM, SAM-MED2D, and SAM-MED3D, on the task of segmenting the aorta and bladder in medical images. It showcases the ground truth segmentations alongside the results produced by each method, highlighting the differences in accuracy and precision. The results suggest that SegVol offers superior performance in both aorta and bladder segmentation tasks compared to the other tested methods. The use of a combination of text and bounding box prompts in SegVol is likely a major contributing factor to its improved performance.

This figure compares the segmentation results of SegVol with other SAM-like interactive methods on the Gall bladder and Left Kidney. It shows the ground truth segmentations alongside the results from MedSAM, SAM (using bounding boxes and points), SAM-MED2D, SAM-MED3D, and SegVol (using text and bounding boxes). The figure visually demonstrates the relative performance of each method for segmenting these anatomical structures.

This figure illustrates the architecture of the SegVol model, which performs 3D medical image segmentation using various types of user prompts (point, bounding box, and text). It highlights the zoom-out-zoom-in mechanism employed for efficient and precise inference. Initially, a rough prediction mask is generated using zoom-out inference, followed by refinement using zoom-in inference on the region of interest (ROI).
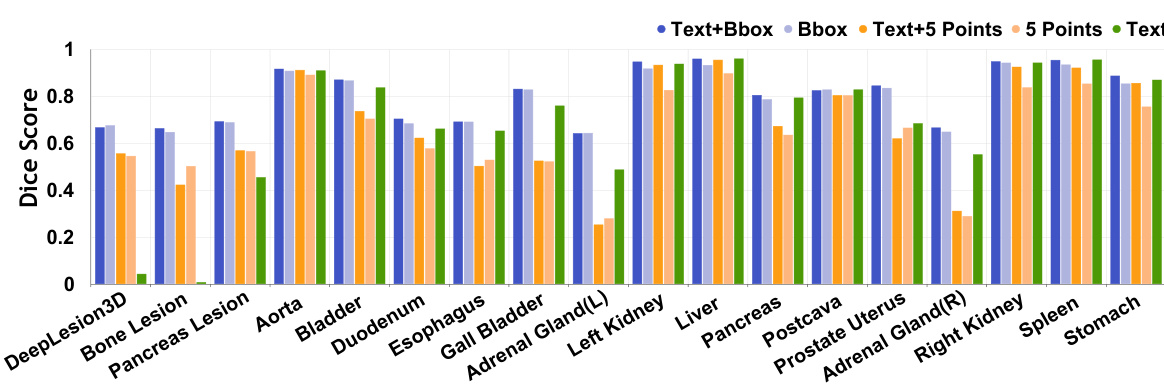
Violin plots showing a comparison of the Dice scores achieved by SegVol and five other SAM-like interactive segmentation methods across various anatomical structures. SegVol demonstrates superior performance in most cases.

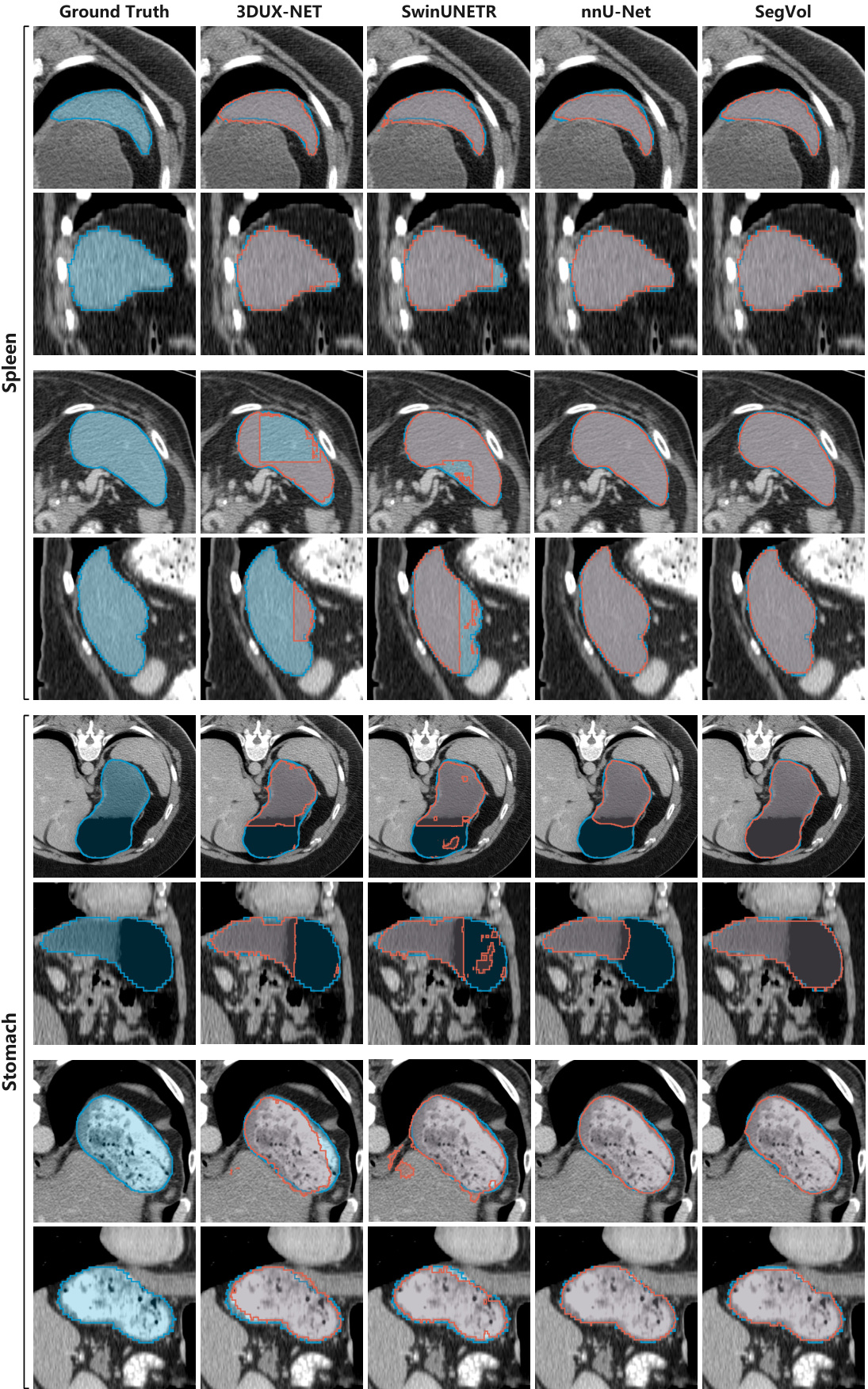
This figure shows the results of applying the SegVol model to unseen MRI data from the CHAOS dataset. The model successfully segments liver, spleen, and kidneys, demonstrating its ability to generalize to different imaging modalities. The comparison between the ground truth segmentations and the SegVol predictions shows the model’s accuracy in identifying organ boundaries.
More on tables
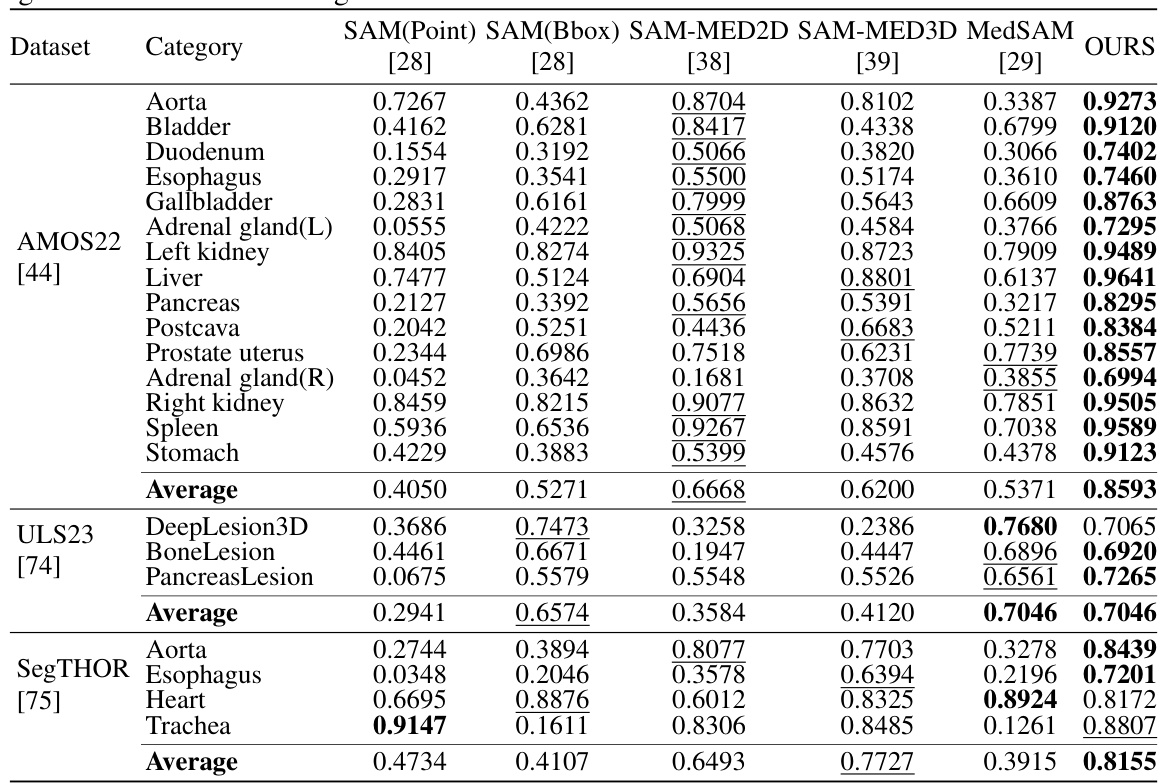
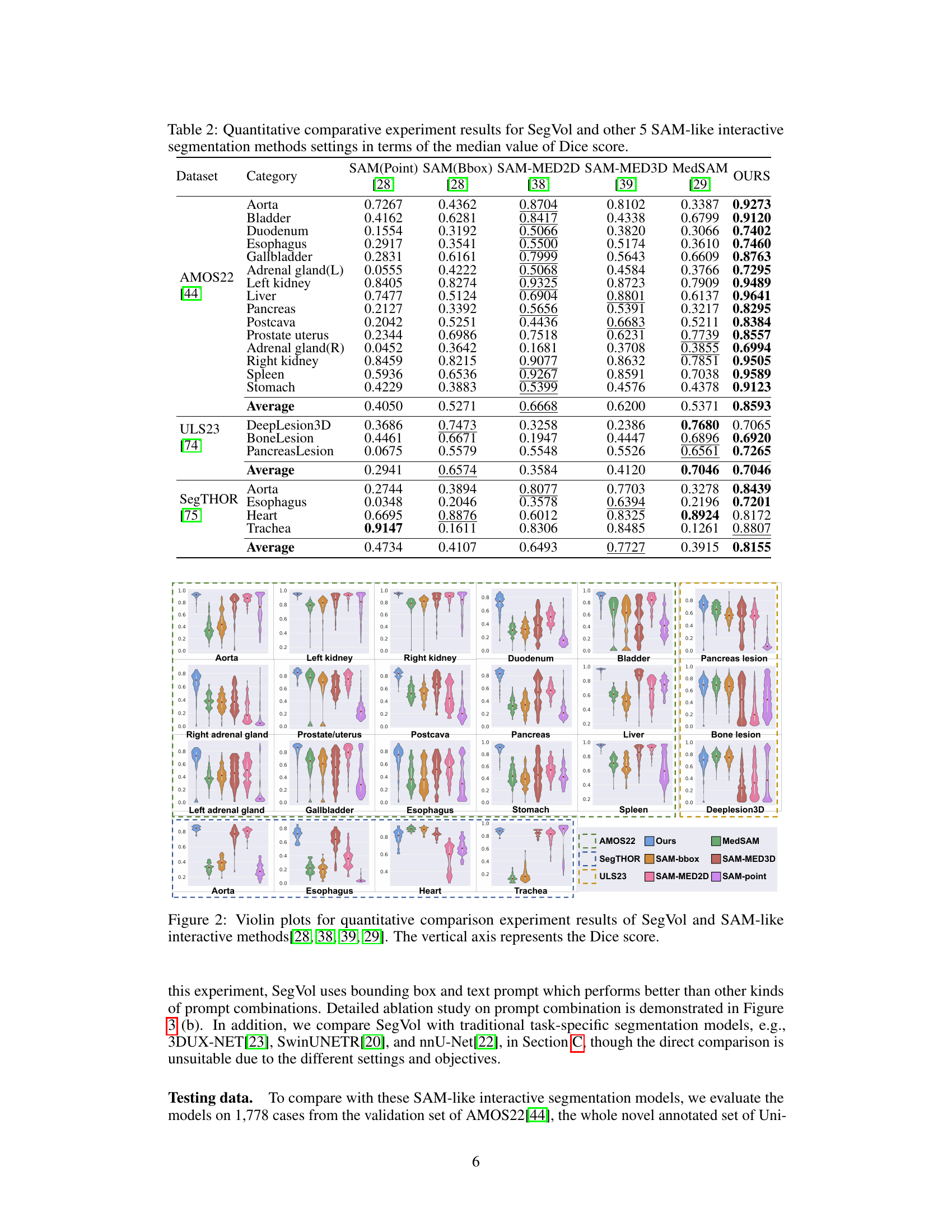
This table presents a quantitative comparison of SegVol’s performance against five other similar interactive segmentation methods across various anatomical structures and datasets. The comparison is based on the median Dice score, a common metric for evaluating segmentation accuracy. Lower Dice scores indicate less accurate segmentations. The table highlights SegVol’s superior performance compared to the other methods across multiple datasets and anatomical categories.
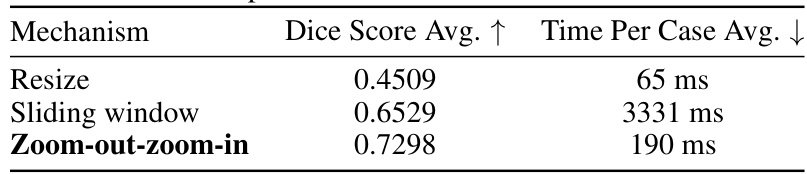
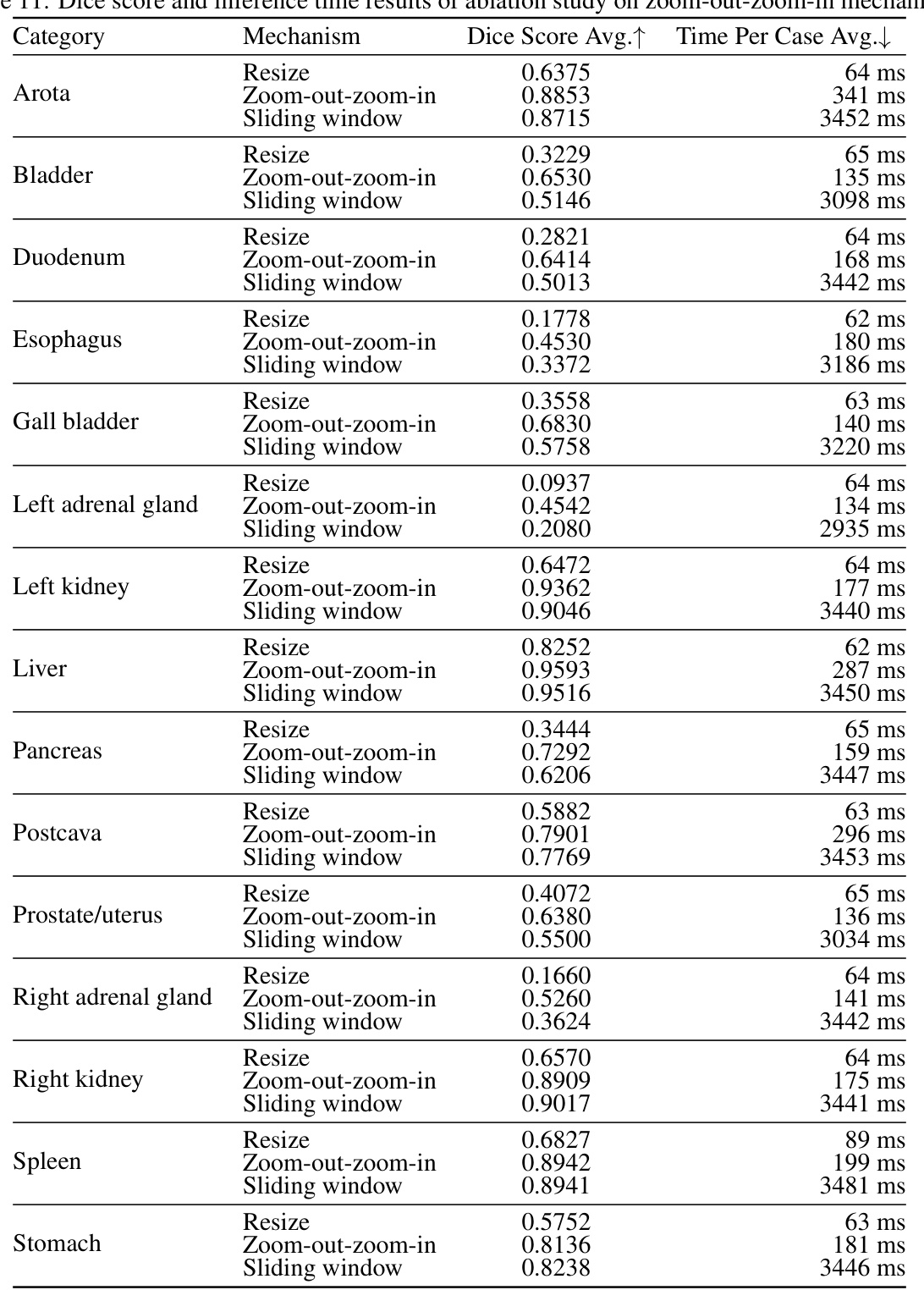
This table presents the ablation study results on the zoom-out-zoom-in mechanism. Three different mechanisms are compared: Resize, Sliding window, and Zoom-out-zoom-in. The results are shown in terms of average Dice score and average time per case. The zoom-out-zoom-in mechanism is shown to achieve the best performance with a significant improvement in the Dice score compared to the other methods.
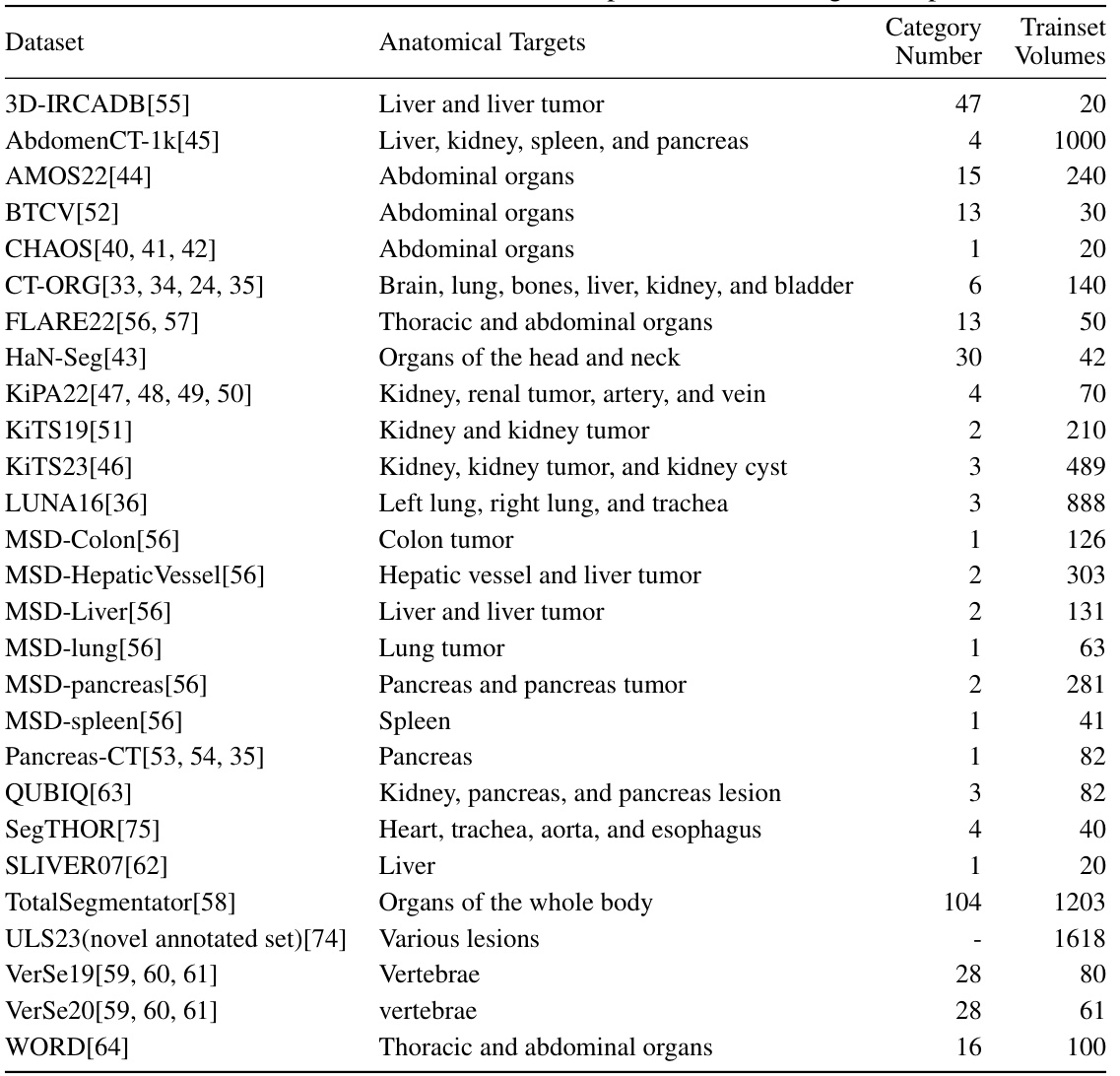
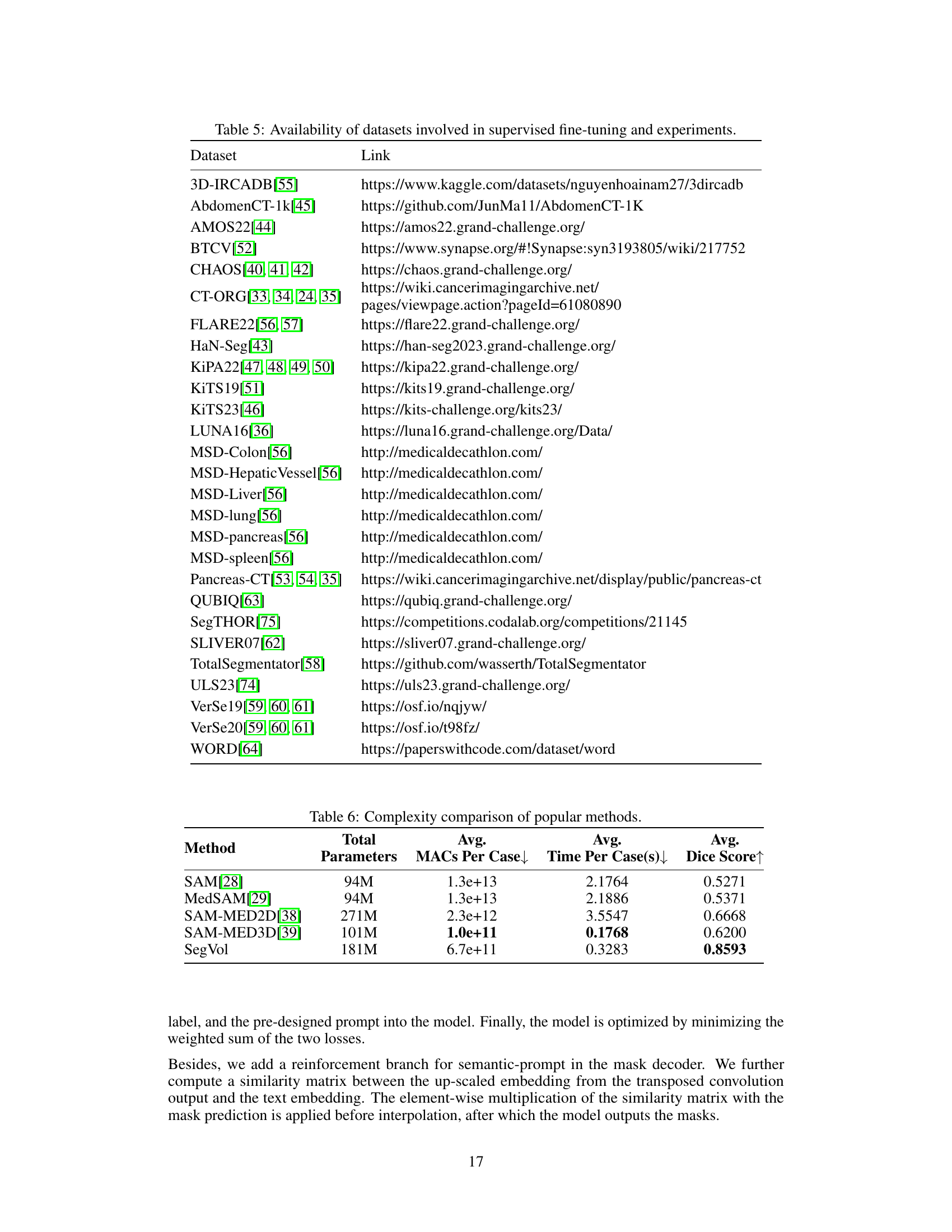
This table lists 25 datasets used for training and evaluating the SegVol model. Each dataset is identified, along with the anatomical targets it contains (organs or tissues), the number of categories present, and the number of training volumes available for each dataset. The datasets cover various body regions and include both organs and lesions.

This table provides links to the 25 open-source datasets used in the paper for supervised fine-tuning and external datasets used in comparative experiments. The table cross-references dataset names with their respective URLs, facilitating access to the data used in the study. This is essential for reproducibility of results.

This table compares the model complexity of SegVol and other SAM-like interactive medical image segmentation methods in terms of the total number of parameters, average MACs (Multiply-Accumulates) per case, average inference time per case, and average Dice score. It highlights the trade-off between model size and performance, showing that SegVol achieves superior performance with a reasonable increase in complexity compared to other methods.

This table compares the average Dice scores achieved by SegVol and nnU-Net on three lesion segmentation tasks: Lung Tumor, Colon Cancer, and Liver Tumor. The Dice score is a common metric for evaluating the accuracy of image segmentation. Higher Dice scores indicate better segmentation performance. The table shows that SegVol significantly outperforms nnU-Net on all three tasks, indicating its superior performance in lesion segmentation.

This table compares the average Dice scores achieved by SegVol and nnU-Net across three lesion segmentation tasks: lung tumor, colon cancer, and liver tumor. The Dice score is a common metric for evaluating image segmentation performance. Higher Dice scores indicate better agreement between the model’s predictions and the ground truth.
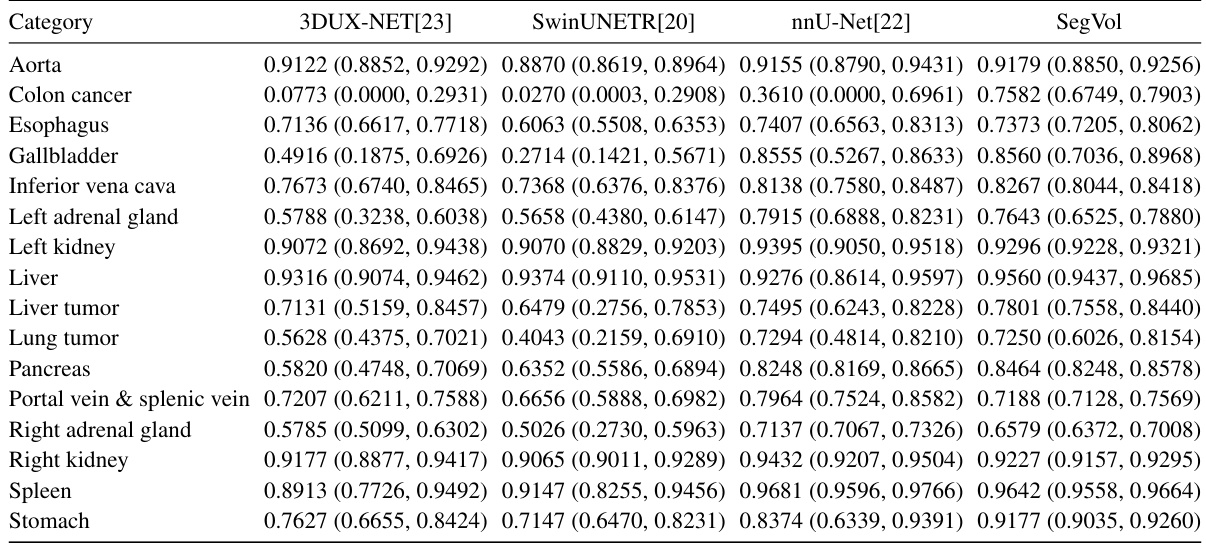
This table presents a comparison of the performance of four different medical image segmentation models (3DUX-NET, SwinUNETR, nnU-Net, and SegVol) on a test set of datasets. The models were all fine-tuned on supervised datasets. The Dice score, a common metric for evaluating segmentation accuracy, is reported for each model and each anatomical structure. The results are presented as median values along with the first and third quartiles to show the distribution of the results.

This table presents a comparison of several interactive segmentation methods, including SegVol, across three different datasets (AMOS22, ULS23, and SegTHOR). The performance is measured using the median Dice score, with first and third quartiles provided to show variability. Each dataset focuses on different anatomical structures, allowing for a comprehensive evaluation across diverse segmentation tasks and datasets.
This table compares several state-of-the-art interactive segmentation methods similar to Segment Anything (SAM). It highlights key differences in their input image domain (2D or 3D), training methods, prompt types supported (points, bounding boxes, or text), and the resolution of the input used during inference. This helps to contextualize the proposed SegVol method within the existing landscape of interactive segmentation techniques.
Full paper#