↗ OpenReview ↗ NeurIPS Proc. ↗ Chat
TL;DR#
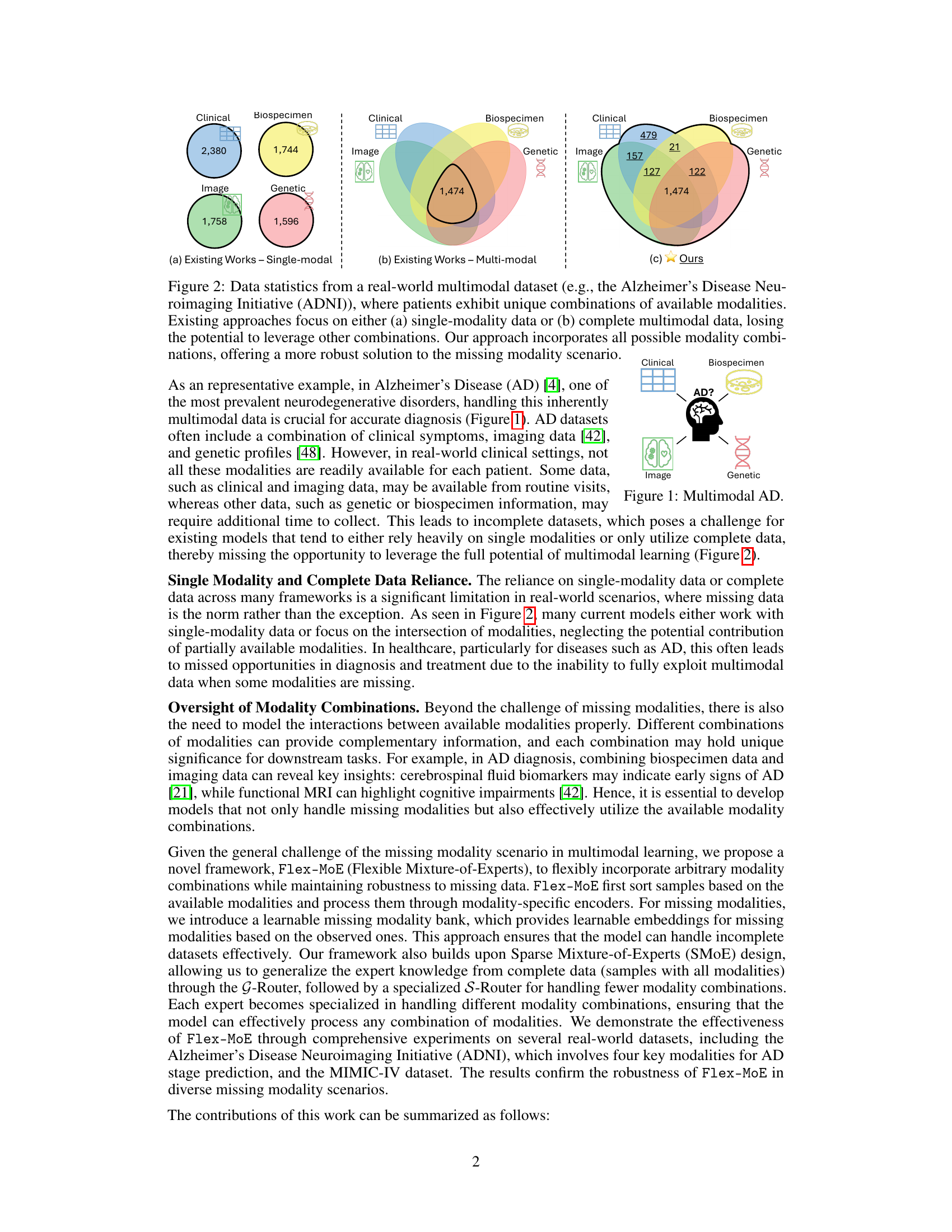
Many real-world applications involve multimodal data, but often some data is missing for certain instances. Existing multimodal learning methods struggle to handle arbitrary combinations of modalities, especially when data is missing. This limits their applicability and necessitates robust solutions.
Flex-MoE, a novel framework, directly addresses this issue. It incorporates a “missing modality bank” to handle missing data and a specially designed Sparse Mixture-of-Experts (MoE) architecture to efficiently manage various modality combinations. Evaluations on Alzheimer’s Disease Neuroimaging Initiative (ADNI) and MIMIC-IV datasets show that Flex-MoE outperforms existing approaches, demonstrating its effectiveness and adaptability in diverse missing modality scenarios.
Key Takeaways#
Why does it matter?#
This paper is crucial for researchers dealing with multimodal data and missing modalities. It offers a novel framework that enhances model robustness and applicability in real-world scenarios, opening new avenues for research and development in various fields. The flexible approach is particularly relevant to applications dealing with incomplete datasets, which are common in healthcare, language, and vision tasks. The superior performance demonstrated on ADNI and MIMIC-IV datasets showcases the efficacy of the proposed method.
Visual Insights#
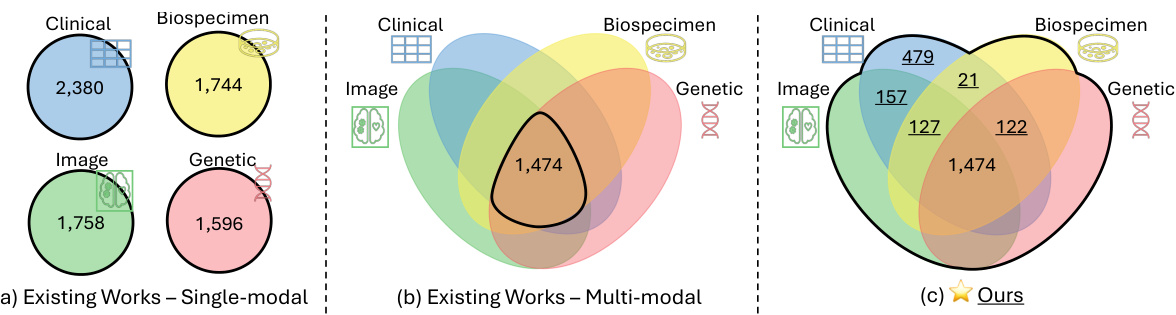
This figure uses the ADNI dataset as an example to illustrate the limitations of existing multimodal learning approaches and highlights the advantages of the proposed Flex-MoE method. Panel (a) shows that existing methods often rely on single-modality data, while panel (b) demonstrates the reliance on complete multimodal data, ignoring numerous combinations with missing modalities. In contrast, panel (c) shows that the proposed method utilizes all possible combinations, handling arbitrary modality combinations robustly.
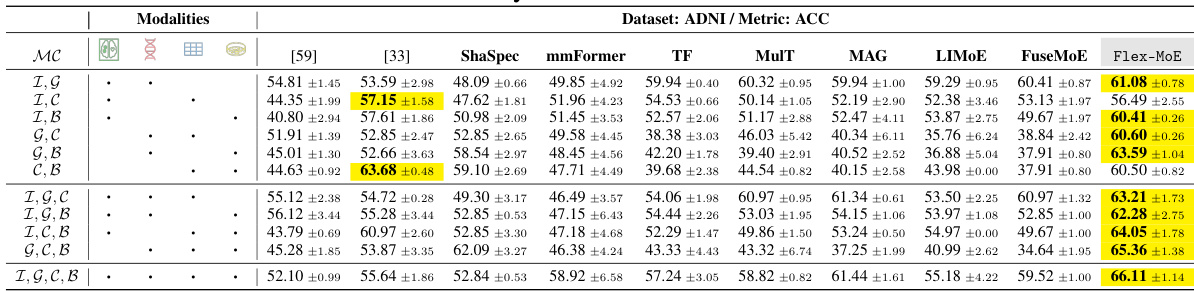
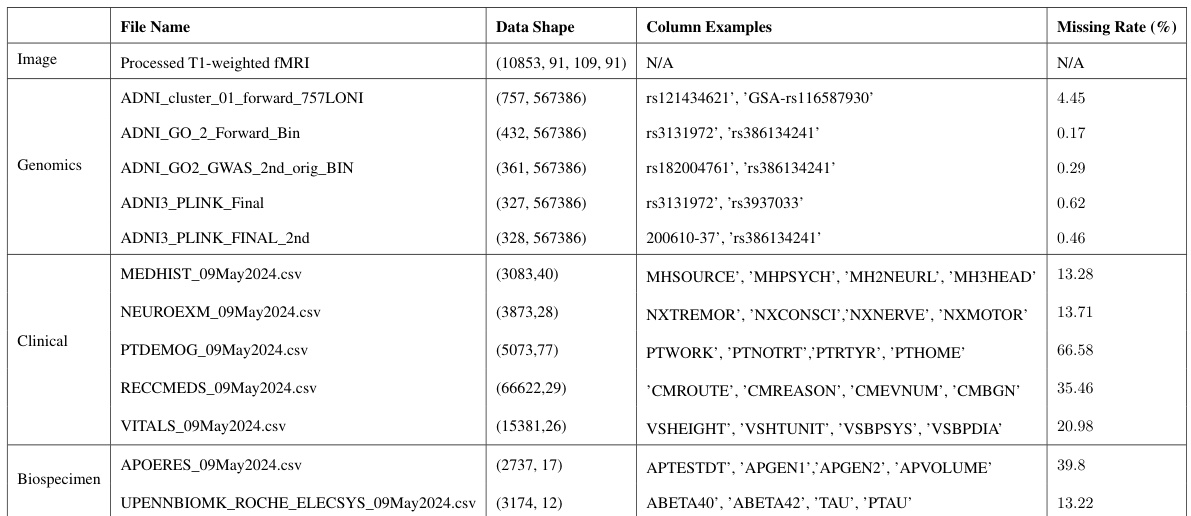
This table presents the performance comparison of various models on the ADNI dataset using the Accuracy (ACC) metric. Different models and varying combinations of modalities (Image, Genetic, Clinical, Biospecimen) are evaluated, showing the impact of modality availability on model accuracy. The table helps to demonstrate Flex-MoE’s performance relative to existing state-of-the-art methods, across different scenarios of missing data.
In-depth insights#
Flex-MoE Overview#
Flex-MoE is a novel framework designed for flexible multimodal learning, addressing the challenge of missing modalities. Its core innovation lies in handling arbitrary modality combinations by employing a missing modality bank to generate embeddings for absent modalities based on observed ones. This is complemented by a sparse Mixture-of-Experts (MoE) architecture. The model cleverly uses a generalized router (G-Router) to train experts on complete data, injecting generalized knowledge. Subsequently, a specialized router (S-Router) assigns experts to handle specific modality combinations present in the input, thus facilitating specialization. This design enables Flex-MoE to effectively leverage all available modality combinations, maintaining robustness even with incomplete data. The overall approach is demonstrated to be highly effective through experiments on real-world datasets like ADNI and MIMIC-IV, showcasing its ability to outperform existing methods in scenarios with missing data.
Modality Handling#
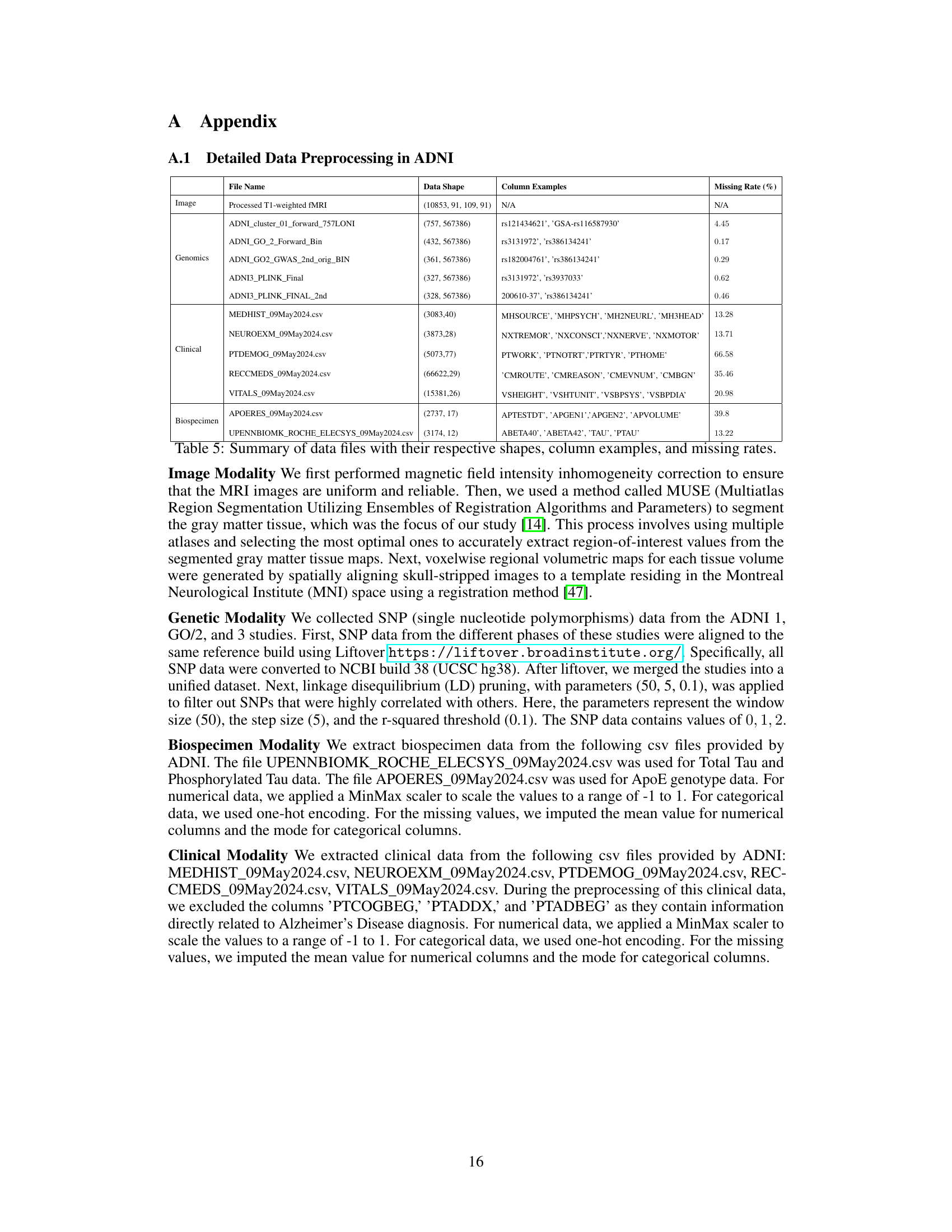
The paper introduces a novel approach to handle missing modalities in multimodal learning. Flex-MoE addresses the challenge of arbitrary modality combinations by employing a ‘missing modality bank’. This bank learns embeddings for missing modalities based on observed combinations, preventing reliance on imputation or zero-padding. The framework uses a unique Sparse Mixture-of-Experts (MoE) design with a generalized router (G-Router) for knowledge sharing across all modalities and a specialized router (S-Router) to assign tasks to experts based on the available modality combination. This flexible approach allows the model to effectively utilize all available information, regardless of data completeness, making it robust in real-world scenarios. The emphasis on handling diverse combinations of modalities is a key strength, moving beyond the typical reliance on single or complete data sets. The results show that Flex-MoE outperforms existing single and multi-modal approaches on the ADNI and MIMIC-IV datasets, showcasing the effectiveness of the missing modality bank and the flexible MoE structure in improving model performance. The work addresses a significant limitation in current multimodal learning techniques, thereby advancing the field’s capability to handle realistic, incomplete datasets.
Missing Data#
The pervasive challenge of missing data significantly impacts the reliability and generalizability of multimodal learning models. The paper highlights how existing frameworks often struggle to handle arbitrary modality combinations, frequently relying on either single modalities or complete datasets, thus neglecting the potential richness of partial data. This limitation is particularly acute in real-world applications, where data scarcity and inconsistencies are commonplace. Addressing missing data requires a flexible approach capable of effectively integrating diverse combinations of available modalities while maintaining robustness to missing information. This necessitates innovative strategies, such as the proposed missing modality bank, to appropriately address the challenge of partial data and leverage all available information for more accurate and robust model training. Flexible Mixture-of-Experts (Flex-MoE) offers a promising solution by incorporating a learnable missing modality bank, allowing the model to effectively generate representations for missing modalities based on observed combinations, thus reducing reliance on imputation or complete data.
Experiment Results#
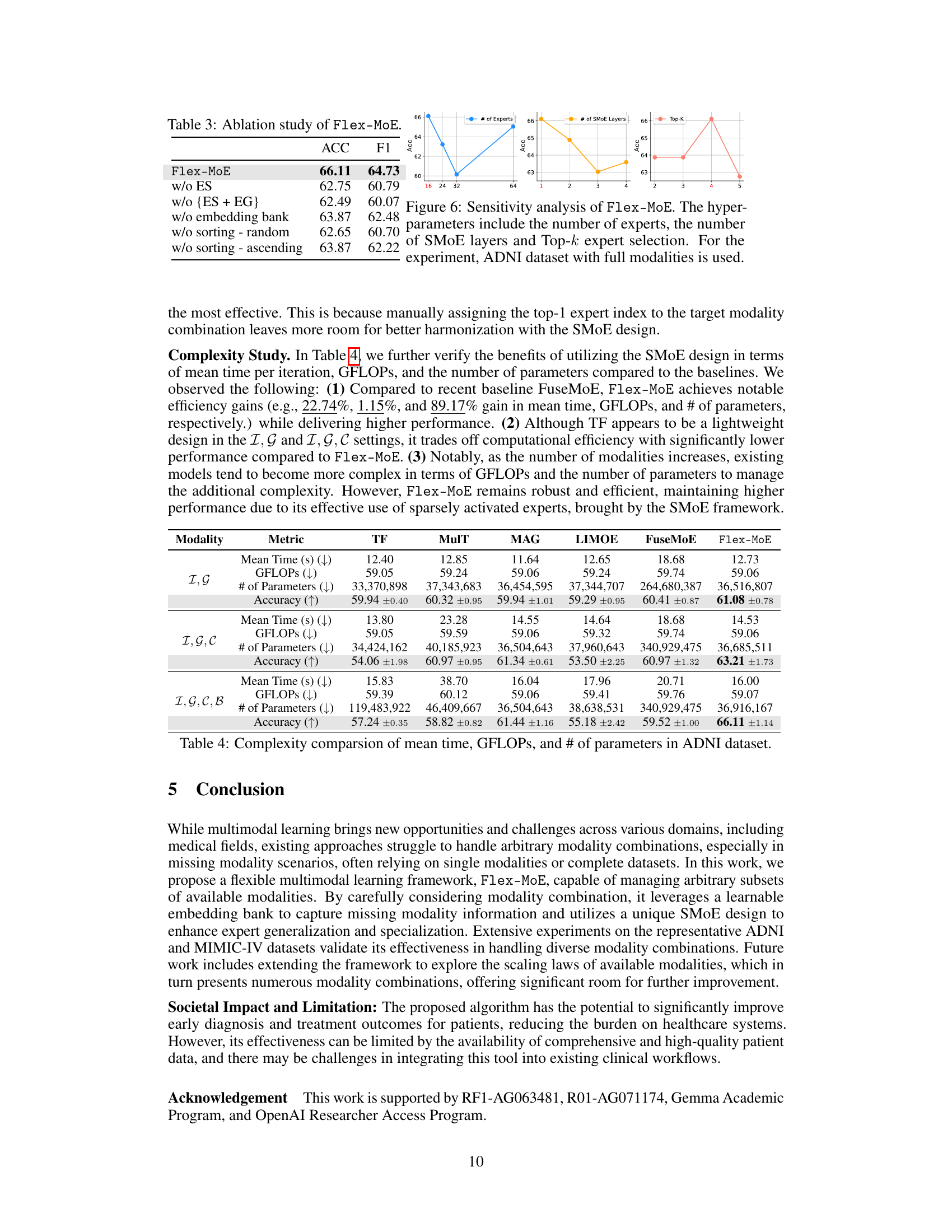
The experimental results section of a research paper is critical; it validates the claims made and demonstrates the effectiveness of proposed methods. A strong results section will clearly present key findings using appropriate metrics, emphasizing statistical significance. Visualizations like graphs and tables should effectively communicate results, making trends and comparisons easily discernible. The discussion should compare the proposed approach against relevant baselines and thoroughly analyze its strengths and limitations, highlighting any unexpected findings or limitations. A robust results section builds confidence in the work’s validity and contribution. Transparency is vital, clearly reporting any experimental details, including the choice of evaluation metrics, data splits, and handling of missing data, enhances the reproducibility of the research and fosters trust within the scientific community. A well-written results section, therefore, should be clear, comprehensive, insightful, and reproducible.
Future Work#
Future research directions stemming from this work on flexible mixture-of-experts (Flex-MoE) for multimodal learning could explore several promising avenues. Scaling the model to handle an even larger number of modalities is crucial, as real-world datasets often contain numerous data sources. This would necessitate further investigation into efficient routing mechanisms within the MoE framework and the management of high-dimensional modality embeddings. Developing more sophisticated missing modality imputation techniques is another key area, potentially leveraging advanced methods like variational autoencoders or generative adversarial networks to produce more realistic and informative missing data representations. Incorporating temporal dependencies within the multimodal data is also important, expanding upon the current static approach by modeling sequences of observations over time. Finally, further investigation into the theoretical properties of Flex-MoE, such as convergence guarantees and generalization bounds, would enhance its robustness and provide a deeper understanding of its behavior in complex scenarios.
More visual insights#
More on figures

This figure illustrates the multimodal nature of Alzheimer’s Disease (AD). It shows that diagnosis of AD often involves integrating information from various sources, including clinical records (symptoms), imaging data (MRI scans, PET scans), genetic profiles, and biospecimens (blood, urine, cerebrospinal fluid). The challenge is that not all of these modalities are always available for each patient, making it difficult for existing models to accurately predict AD stages.
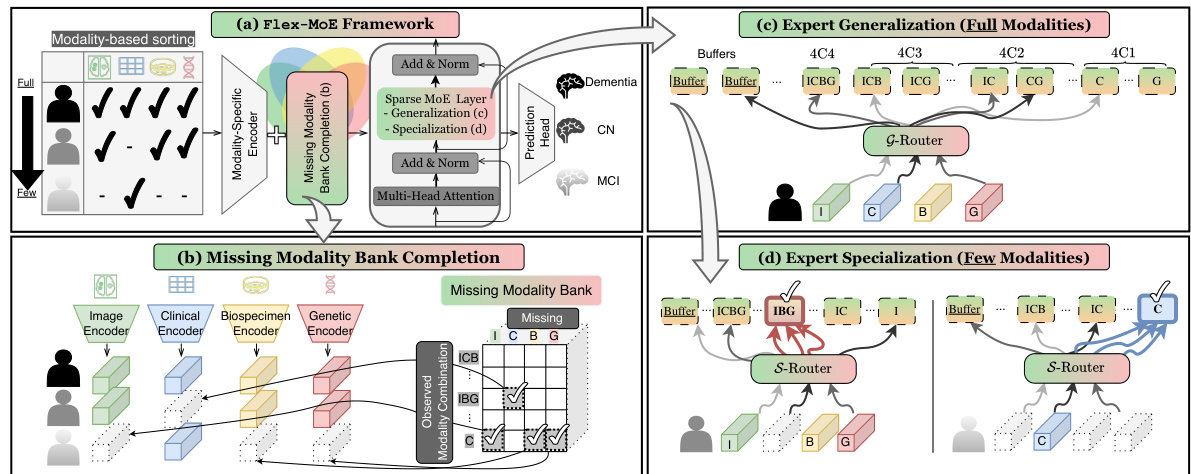
This figure illustrates the Flex-MoE framework, showing the process of sorting samples by modality availability, handling missing modalities using a missing modality bank, and employing a Sparse Mixture-of-Experts (SMoE) layer with both generalized and specialized experts. The G-Router handles samples with full modalities, while the S-Router specializes in handling samples with fewer modalities.

This figure shows a comprehensive illustration of the Flex-MoE model’s architecture. It details the process of handling missing modalities using a missing modality bank and using a two-stage routing mechanism (G-Router and S-Router) for training both generalized and specialized experts to handle various modality combinations.

This figure visualizes the activation ratio of input modality combinations across different expert indices in the Flex-MoE model. It demonstrates how the model utilizes both generalized knowledge from samples with complete modalities and specialized knowledge from samples with fewer modalities, leading to effective handling of various modality combinations.
More on tables
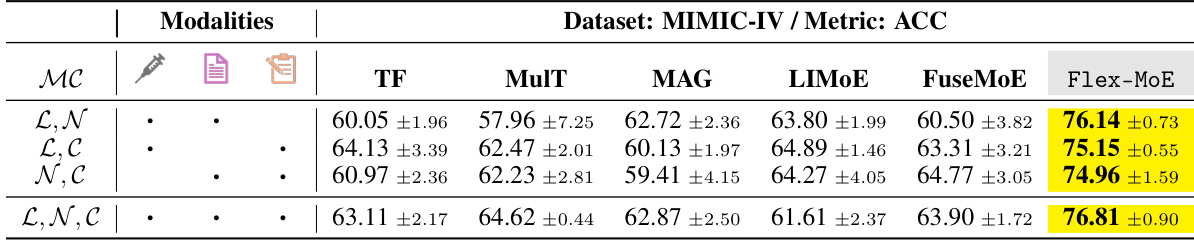
This table presents the performance of various models on the MIMIC-IV dataset using the accuracy (ACC) metric. The models are evaluated under different modality combinations, specifically those including Lab and Vital values, Clinical Notes, and ICD-9 codes. The table shows the accuracy of each model for each combination, highlighting the performance of the proposed Flex-MoE model compared to existing state-of-the-art methods.

This table presents the results of ablation experiments conducted on the Flex-MoE model. By systematically removing components of the model (expert specialization, expert generalization, embedding bank, and sorting), the impact on the model’s performance (measured by Accuracy and F1 score) is evaluated. This helps to understand the contribution of each component to the overall performance of Flex-MoE.

This table presents the performance comparison of various models (Flex-MoE and several baselines) on the ADNI dataset in terms of accuracy (ACC). The models were tested with different combinations of modalities (Image, Genetic, Clinical, Biospecimen), and the results highlight Flex-MoE’s superior performance in handling missing modalities and arbitrary modality combinations. The MC column indicates the specific observed modality combination for each row.
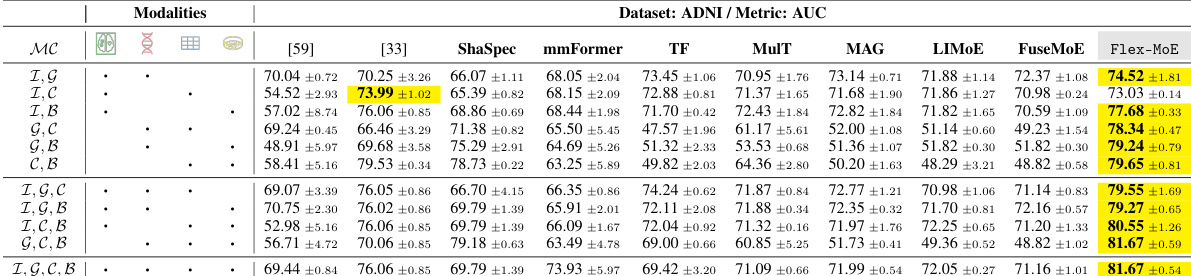
This table presents the performance comparison of various models on the ADNI dataset, using the Accuracy (ACC) metric. It shows how different models perform across different combinations of four modalities (Image, Genetic, Clinical, Biospecimen). The table enables a comparison of single-modality and multi-modality approaches, as well as different state-of-the-art methods, including the proposed Flex-MoE model. Each row represents a specific combination of available modalities, and the columns indicate different models’ ACC values.

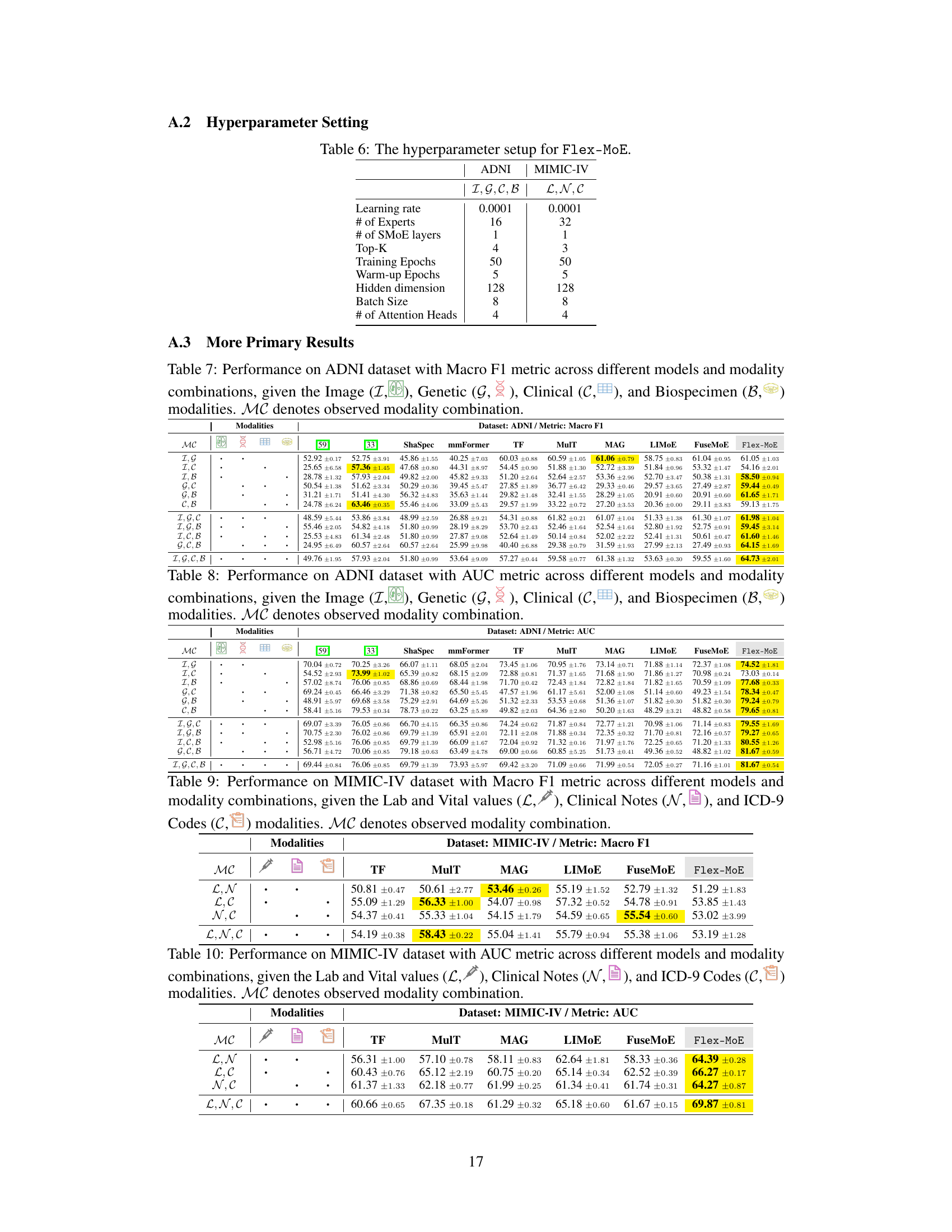
This table shows the hyperparameter settings used for training the Flex-MoE model on two different datasets: ADNI and MIMIC-IV. For each dataset, the table specifies the learning rate, the number of experts, the number of SMoE layers, the top-K value for expert selection, the number of training epochs, the number of warm-up epochs, the hidden dimension of the model, the batch size, and the number of attention heads.

This table shows a comparison of the performance (Accuracy) of different models on the ADNI dataset for Alzheimer’s Disease prediction. The models are tested across various combinations of available modalities (Image, Genetic, Clinical, and Biospecimen). The table highlights the superior performance of Flex-MoE across different modality scenarios.
This table presents the performance comparison of various models on the ADNI dataset using the accuracy (ACC) metric. It shows the performance for different combinations of modalities (Image, Genetic, Clinical, and Biospecimen) with various missing data scenarios represented by ‘MC’. The results highlight the performance of Flex-MoE compared to existing single and multi-modal methods.

This table presents the performance comparison of different models on the MIMIC-IV dataset using the accuracy (ACC) metric. It shows results for various combinations of available modalities (Lab and Vital values, Clinical Notes, and ICD-9 Codes), demonstrating how each model handles different levels of missing data. Flex-MoE’s performance is compared against several baseline methods, highlighting its effectiveness in various missing-modality scenarios.

This table presents the performance comparison of various models on the MIMIC-IV dataset using the AUC metric. Different models, including TF, MulT, MAG, LIMOE, FuseMoE, and Flex-MoE, are compared under various modality combinations (Lab and Vital values, Clinical Notes, and ICD-9 Codes). The results demonstrate the effectiveness of Flex-MoE in handling different combinations of modalities.
Full paper#